Genomic and epidemiological features of two dominant methicillin-susceptible Staphylococcus aureus clones from a neonatal intensive care unit surveillance effort
Abstract
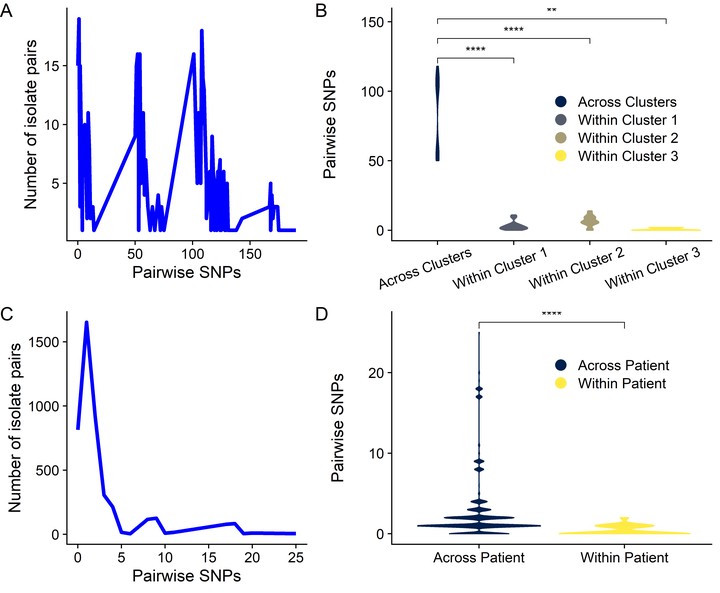
Methicillin-susceptible Staphylococcus aureus (MSSA) is a more prevalent neonatal intensive care unit (NICU) pathogen than methicillin-resistant S. aureus (MRSA). However, the introduction and spread of MSSA, the role of systematic decolonization, and optimal infection prevention and control strategies remain incompletely understood. We previously screened infants hospitalized in a university-affiliated level III to IV NICU twice monthly over 18 months for S. aureus colonization and identified several prevalent staphylococcal protein A (spa) types. Here, we performed whole-genome sequencing (WGS) and phylogenetic comparisons of 140 isolates from predominant spa types t279, t1451, and t571 to examine possible transmission routes and identify genomic and epidemiologic features associated with the spread of dominant clones. We identified two major MSSA clones: sequence type 398 (ST398), common in the local community, and ST1898, not previously encountered in the region. ST398 NICU isolates formed distinct clusters with closely related community isolates from previously published data sets, suggesting multiple sources of acquisition, such as family members or staff, including residents of the local community. In contrast, ST1898 isolates were nearly identical, pointing to clonal expansion within the NICU. Almost all ST1898 isolates harbored plasmids encoding mupirocin resistance (mupA), suggesting an association between the proliferation of this clone and decolonization efforts with mupirocin. Comparative genomics indicated genotype-specific pathways of introduction and spread of MSSA via community-associated (ST398) or health care-associated (ST1898) sources and the potential role of mupirocin resistance in dissemination of ST1898. Future surveillance efforts could benefit from routine genotyping to inform clone-specific infection prevention strategies.
Importance
Methicillin-susceptible Staphylococcus aureus (MSSA) is a significant pathogen in neonates. However, surveillance efforts in neonatal intensive care units (NICUs) have focused primarily on methicillin-resistant S. aureus (MRSA), limiting our understanding of colonizing and infectious MSSA clones which are prevalent in the NICU. Here, we identify two dominant colonizing MSSA clones during an 18-month surveillance effort in a level III to IV NICU, ST398 and ST1898. Using genomic surveillance and phylogenetic analysis, coupled with epidemiological investigation, we found that these two sequence types had distinct modes of spread, namely the suggested exchange with community reservoirs for ST398 and the contribution of antibiotic resistance to dissemination of ST1898 in the health care setting. This study highlights the additional benefits of whole-genome surveillance for colonizing pathogens, beyond routine species identification and genotyping, to inform targeted infection prevention strategies.